
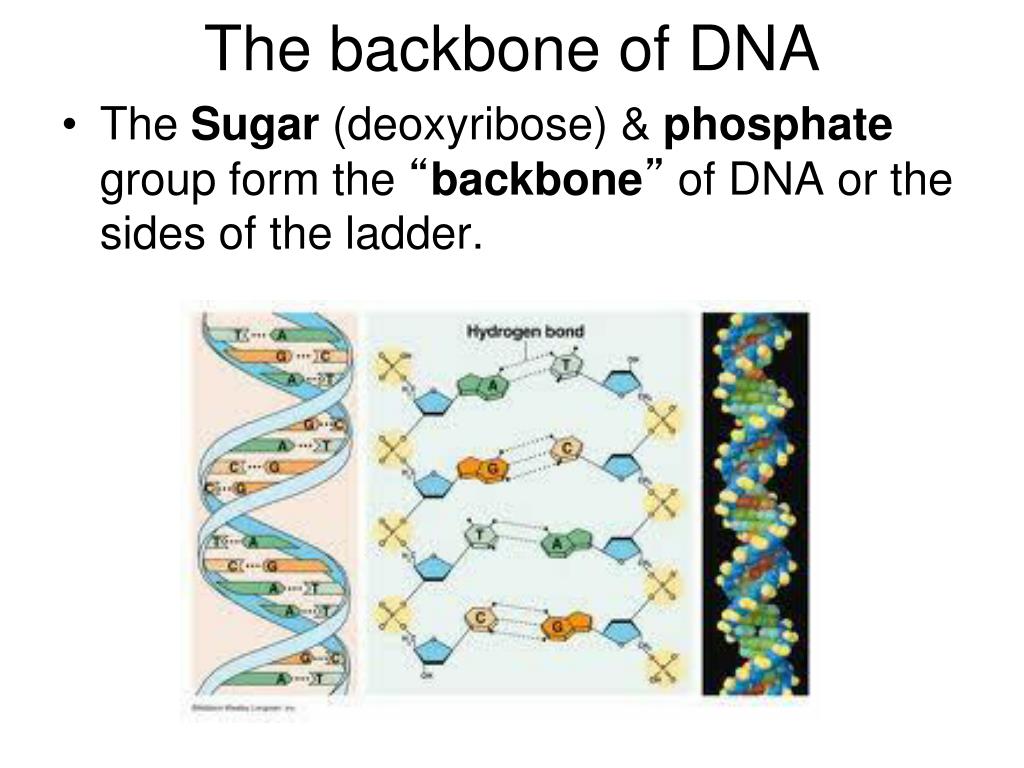
The base is attached to the ribose's 1′ position, and the phosphate is attached to the 5′ position. Carbon residues in the pentose are numbered 1′ through 5′ (the prime distinguishes these residues from those in the base, which are numbered without using a prime notation).
Each nitrogenous base in a nucleotide is attached to a sugar molecule, which is attached to one or more phosphate groups.įigure 3.31 Three components comprise a nucleotide: a nitrogenous base, a pentose sugar, and one or more phosphate groups. Three components comprise each nucleotide: a nitrogenous base, a pentose (five-carbon) sugar, and a phosphate group ( Figure 3.31). The nucleotides combine with each other to form a polynucleotide, DNA or RNA. Other types of RNA-like rRNA, tRNA, and microRNA-are involved in protein synthesis and its regulation.ĭNA and RNA are comprised of monomers that scientists call nucleotides. This intermediary is the messenger RNA (mRNA). The DNA molecules never leave the nucleus but instead use an intermediary to communicate with the rest of the cell. The other type of nucleic acid, RNA, is mostly involved in protein synthesis. DNA controls all of the cellular activities by turning the genes “on” or “off.” Many genes contain the information to make protein products. A chromosome may contain tens of thousands of genes. In eukaryotic cells but not in prokaryotes, DNA forms a complex with histone proteins to form chromatin, the substance of eukaryotic chromosomes. The cell's entire genetic content is its genome, and the study of genomes is genomics. In prokaryotes, the DNA is not enclosed in a membranous envelope. It is in the nucleus of eukaryotes and in the organelles, chloroplasts, and mitochondria. DNA is the genetic material in all living organisms, ranging from single-celled bacteria to multicellular mammals. The two main types of nucleic acids are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). They carry the cell's genetic blueprint and carry instructions for its functioning. Nucleic acids are the most important macromolecules for the continuity of life.
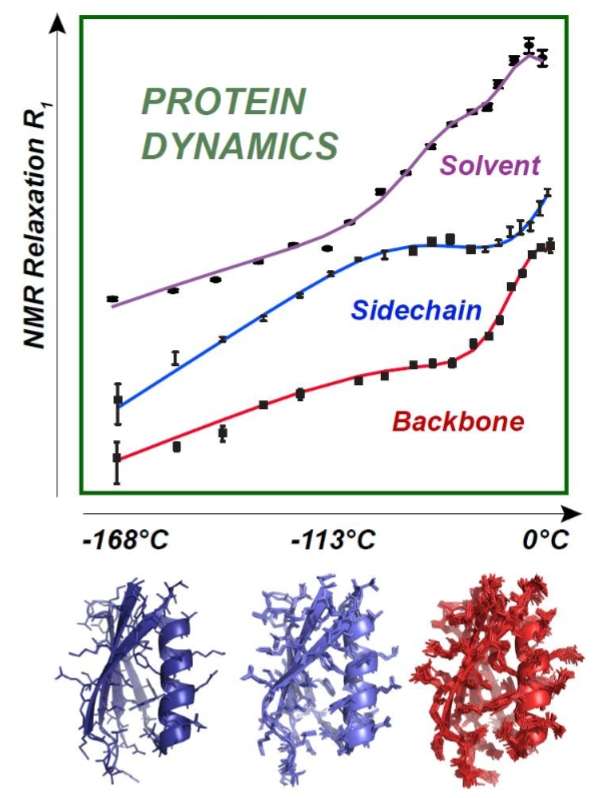
The selected target protein was lysine-, arginine-, ornithine-binding protein (LAO), which comprises two domains and undergoes large domain motions. In the present study, we investigated slow dynamics of the protein backbone using MD simulation and tICA. Some of these motions might occur on a slow time scale: we hypothesize that if so, we could effectively detect and characterize them using tICA. However, the protein changes its conformation not only through domain motions but also by various types of motions involving its backbone and side chains. Our previous study focused on domain motions of the protein and examined its dynamics by using rigid-body domain analysis and tICA. We recently proposed the method of time-structure based independent component analysis (tICA) to examine the slow dynamics involved in conformational fluctuations of a protein as estimated by molecular dynamics (MD) simulation.


 0 kommentar(er)
0 kommentar(er)
